Difference between revisions of "Educational Data Mining"
m (New page: Mastery Grids aims to provide better experience to both students and instructors by using automatic tools and techniques that are derived by machine learning and data mining algorithms on ...) |
m |
||
| Line 8: | Line 8: | ||
Categories | Categories | ||
[[Image:ProblemSolvingGenome.png|650x1300px]] | [[Image:ProblemSolvingGenome.png|650x1300px]] | ||
| + | |||
| + | |||
| + | == Tensor and Matrix Factorization in Predicting Student Performance == | ||
| + | This research aims to predict success and failure of students in future questions using collaborative-filtering approaches including tensor and matrix factorizations. One of the benefits of these models is their ability to estimate the underlying skills of questions automatically. In this work, we research on these approaches on educational data in two settings: considering the attempt (time) sequence of students and considering the underlying concept structure of questions. We compare these approaches with state of the art methods such as Feature-Aware Knowledge Tracing (FAST), Bayesian Knowledge Tracing, and Performance Factor Analysis. | ||
| + | |||
| + | == Feedback-Driven Tensor Factorization or Simultaneously Modeling Student Knowledge and Course Content == | ||
Revision as of 20:18, 4 April 2016
Mastery Grids aims to provide better experience to both students and instructors by using automatic tools and techniques that are derived by machine learning and data mining algorithms on its data. In this section, we introduce some of the projects performed on Mastery Grids' data for this purpose.
Problem Solving Genome for Students
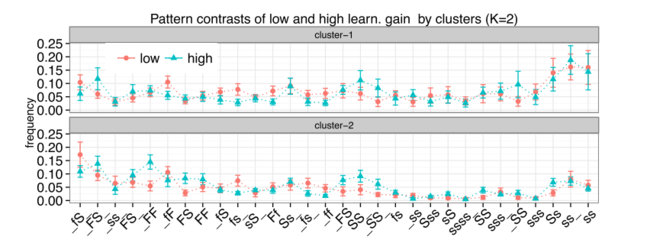
In this research we study the students' behavioral patterns in attempting quizzes and repeating them. We look at attempt sequence of students, model, and examine patterns of student behavior with parameterized exercises. Starting with micro-patterns (genes) that describe small chunks of repetitive behavior of students, we construct individual student profiles (genomes) as frequency profiles. These profiles show the dominance of each gene (repetitive pattern) in individual behavior. We cluster the students using these profiles and study their learning gains in relation to their constructed genome. The exploration of student genomes revealed the individual genome is considerably stable, distinguishing students from their peers. It uniquely identifies a user among other users over the whole duration of the course despite a considerable growth of student knowledge over the course duration. While the problem complexity does affect the behavior patterns as well, we demonstrated that the genome is defined by some inherent characteristics of the user rather than a difficulty profile of the problems she solves. In the group level, all students can be most reliably split into just two cohorts that differ considerably by their behavior. After that split, we are able to contrast successful and less successful learners by their behavior and identify “beneficial” and “harmful” genes for each cohort. In particular, it is interesting to observe that the behavior of successful learners in one cohort is somewhat closer to the behavior of the opposite cohort.
Tensor and Matrix Factorization in Predicting Student Performance
This research aims to predict success and failure of students in future questions using collaborative-filtering approaches including tensor and matrix factorizations. One of the benefits of these models is their ability to estimate the underlying skills of questions automatically. In this work, we research on these approaches on educational data in two settings: considering the attempt (time) sequence of students and considering the underlying concept structure of questions. We compare these approaches with state of the art methods such as Feature-Aware Knowledge Tracing (FAST), Bayesian Knowledge Tracing, and Performance Factor Analysis.